Portal for Marine Ecological GenomiX (MegX)One central theme of the MicroB3 Project is to enable open access and a transparent data flow for marine microbial ecosystems research and biotechnology. This data flow includes contextual data about the environment as well as sequence data from initial sampling to web based end-user access, as well as to various data products derived from the data by the various analysis pipelines. One of the main tangible products of MicroB3 is the Microbial Ecological Genomics Database (MegDB). MegDB includes several extensions which allow efficient integrated access to molecular sequences and environmental data. It is also used in an architecture which bridges the gap between analytical pipelines and storage of data in a scalable manner. MegDB serves as the main data integration platform of the Portal for Marine Ecological GenomiX (MegX), as part of the MicroB3 Information System. MegX is a website for specialized georeferenced databases and tools for the analysis of marine bacterial, archaeal, and phage genomes and metagenomes. |
|
The MicroB3 Project (Microbial Biodiversity, Bioinformatics and Biotechnology) (EU FP7 project)
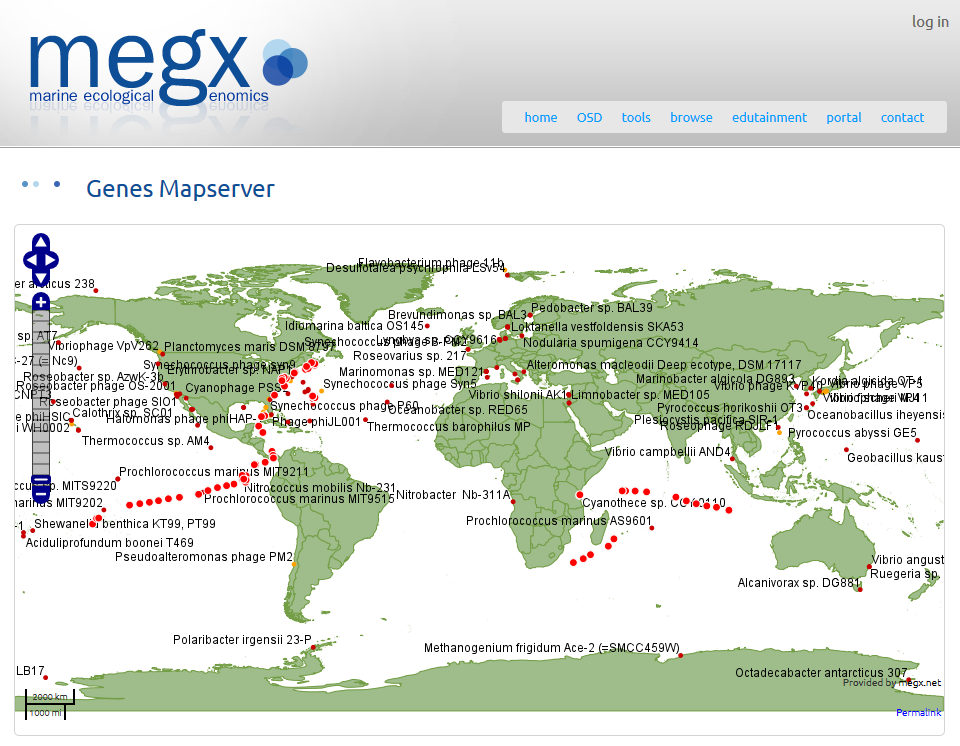
The MegX database, MegDB (Microbial Ecological Genomics DataBase), is a collection of publicly available georeferenced marine bacterial and archaeal genomes and metagenomes, including the Global Ocean Sampling (GOS) reads.
MegX Tools:
MegX also provides access to tools developed by the Microbial Genomics Group at the Max Planck Institute for Marine Microbiology.
- MetaMine: A biologist-centric software tool for data mining and comparison of prokaryotic (meta)genomes.
- MetaLook: A Java-based software tool for marine ecological genomics. It offers a three dimensional environment for visualization and data-mining of DNA sequence information (genomes, metagenomes) in a geographical and ecological context.
- JCoast: A biologist-centric software tool for data mining and comparison of prokaryotic (meta)genomes.
- TETRA: A web-service and a stand-alone program for the analysis and comparison of tetranucleotide usage patterns in DNA sequences.
- MADA: A micro array data analyzing tool, focusing on usability and calculation transparency. It provides a conceptual and well documented step-by-step access to the analyzing process.
- RibAlign: A tool for those interested in phylogenetic reconstructions based on concatenated ribosomal protein sequences of Bacteria. It provides a fast and scalable database that has been specifically adapted to ribosomal protein sequences.
- MicHanThi: Automated prediction of gene functions.
- MetaBar: The tool for Consistent Contextual Data Acquisition.
- TaxSOM: A bioinformatic tool for taxonomic classification of (meta-)genomic DNA-fragments based on taxon-specific DNA-base composition characteristics using Self-Organizing Maps.
- CDinFusion: A submission-preparation-tool for the integration of contextual data (CD) with sequence data.